RNAseq Database Model
RNAseq Database Model
Here you can find a project from the subject Databases & Web Development, where I had to desing an SQL database according to the following requirements:
You are the manager of a bioinformatics support service and need to build a database to manage data from your users' studies. Define a data model (entities, atributes and relationships) to hold data from a series of RNAseq (analysis). Data should include:
- Genes included in the study.
- Reference, suppliers, for sequencing reagents and equipment used.
- Sample and user details.
- Results: genes, expression values, differential expression analysis.
- References.
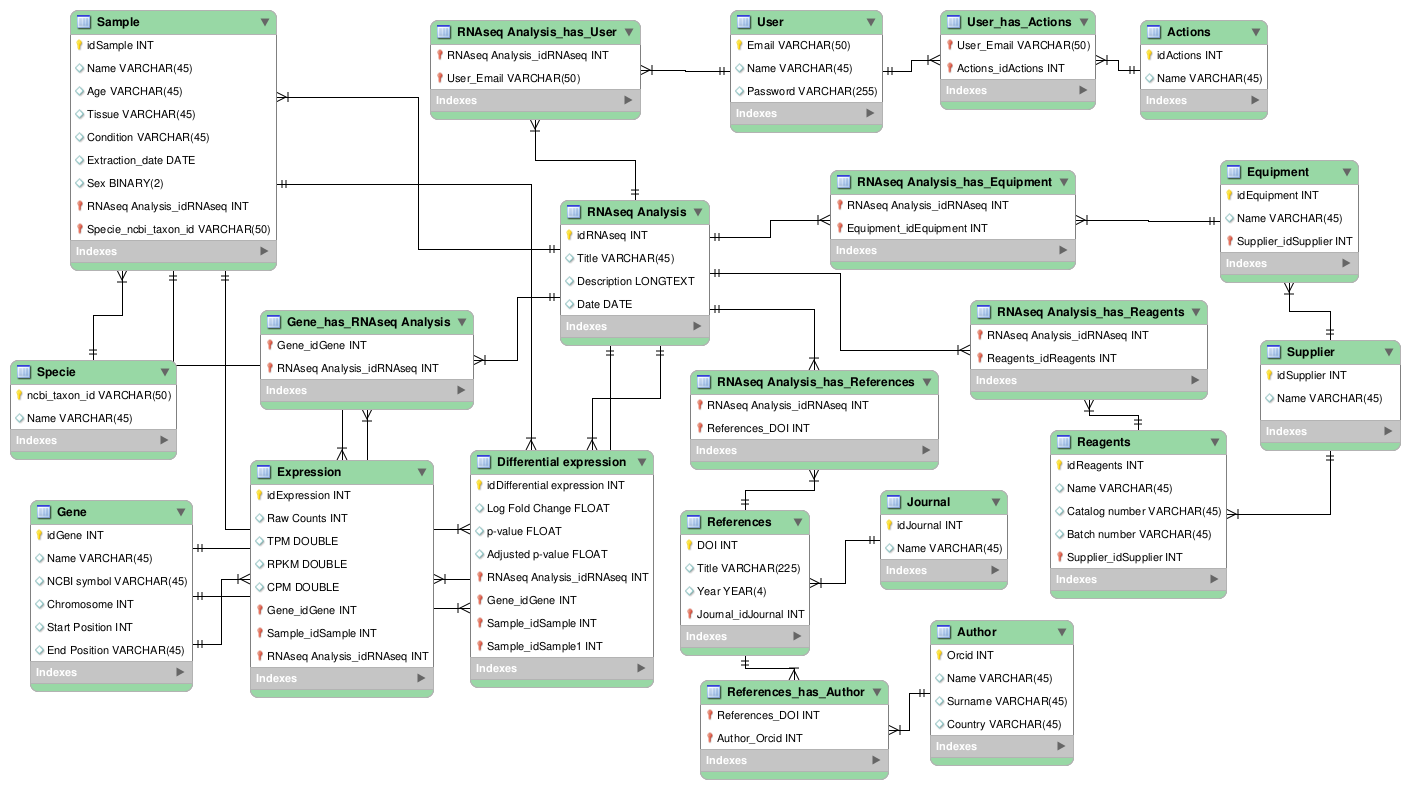
I will try to explain the logic behind my model:
-
The database is centered around the entity RNAseq Analysis which represents a single RNAseq experiment.
-
There are also Users who have access the database. On one hand, different users will have different actions or permissions, for example, in a group all the members may be able to access the data but only the bioinformatician may be able to modify the data. On the other hand, users are connected with RNAseq Analysis in a n:m way.
-
Then we have the entity Sample, which contains all the important information about the origin of the sample (metadata), such as tissue, age, condition or sex. The samples are connected to the RNAseq Analysis entity in a 1:n manner, as we would expect an experiment to have different samples but a sample to appear only on one experiment; and to the Species entity, also in a 1-to-many relation (a sample can only come from one specie, but we could have many samples from the same specie).
-
For the Gene I included attributes I found important, such as the chromosome or the starting and ending postion. I also wanted to include some form of symbol/notation, which raised the problem of having different organisms and thus different gene names (HUGO vs. MGI for example). At the end I chose the NCBI notation, however more thought should be given to this. As for its relations, it has one with RNAseq Analysis in a many-to-many manner, as in one experiment we will look at many genes, and a gene can appear in different experiments. I think it makes sense to unite these two entities together (and not with Species for example), as I would expect all the samples of an experiment to be mapped against the same genome, thus generating the same "instances" of genes.
-
Then we have the Expression, where I included both the raw counts and different normalization methods, like TPM, RPKM or CPM. This may be a bit of an overkill, as they can be computed from the raw counts. The expression has a 1-to-many relationship with the gene, sample and RNAseq analysis entities (one gene, sample or RNAseq experiment will have "generate" expression data but one expression data can only come from one gene, sample and RNAseq experiment).
-
I decided that the Differential Expression should be a separate entity, with attributes LogFold change, p-value and adjusted p-value. As for the relationships it has a 1-to-many relation with the gene and RNAseq analysis. It is also related with the samples two times. This is necessary as to do a differential expression we need to compare two samples/conditions. However, this creates a problem. In most RNAseq experiments there will be multiple replicates of the same condition, which in our model would be classified as different samples, but the differential expression is calculated considering the condition of interest, thus pooling the samples together, which is not possible under the current model. Maybe there should be a new entity, Condition, that would be "in the middle" of Sample and Differential Expression.
-
Then we have the Equipment and Reagents entities, which operate in a similar manner. They have a many-to-many relationship with RNAseq analysis, as one equipment or reagent can be used in many experiment and one experiment will require different equipment and reagents; and a 1-to-many with the entity Supplier.
-
Finally we have the entity Reference, which I interpreted as containing the published articles where one particular RNAseq experiment was used. Thus, it has a many-to-many relationship with the entity RNAseq Analysis, as it may happen that an experiment is discussed in different articles and an article could use or compare different RNAseq analysis. Additionally, it is also related to the entities Author, in a many-to-many manner, and Journal, in a 1-to many manner.
