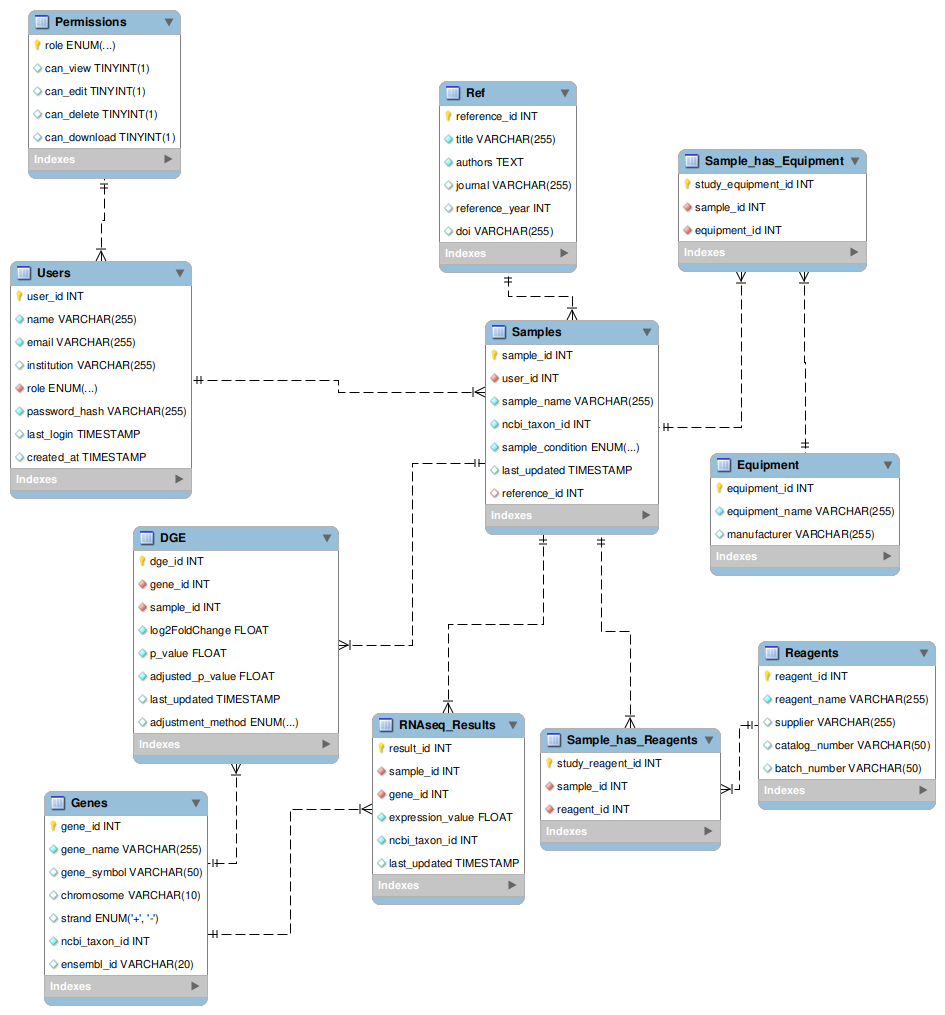
This database model is designed for managing RNAseq data, tracking user studies, sequencing reagents, equipment, and analysis results. It includes the following entities:
- Users and Permissions: Manages researchers and administrators, controlling access levels for viewing, editing, and deleting data.
- Samples: Represents biological samples used in RNAseq studies, linked to users and associated with an NCBI Taxon ID for species classification.
- Genes: Stores gene information, including gene names, symbols, Ensembl IDs, and chromosome locations.
- RNAseq_Results: Contains expression values for each gene-sample pair, tracking measured transcription levels.
- Differential Gene Expression (DGE): Stores statistical results from differential expression analysis, including log2 fold change and p-values.
- Reagents and Equipment: Tracks sequencing reagents, suppliers, and batch numbers, along with the equipment used in sequencing.
- References: Stores literature citations related to studies, linked to samples for research traceability.
- Relationships: Samples are linked to users, genes are referenced in RNAseq results, and reagents/equipment are assigned to specific experiments.
This database structure ensures efficient data organization and traceability for RNA sequencing projects.