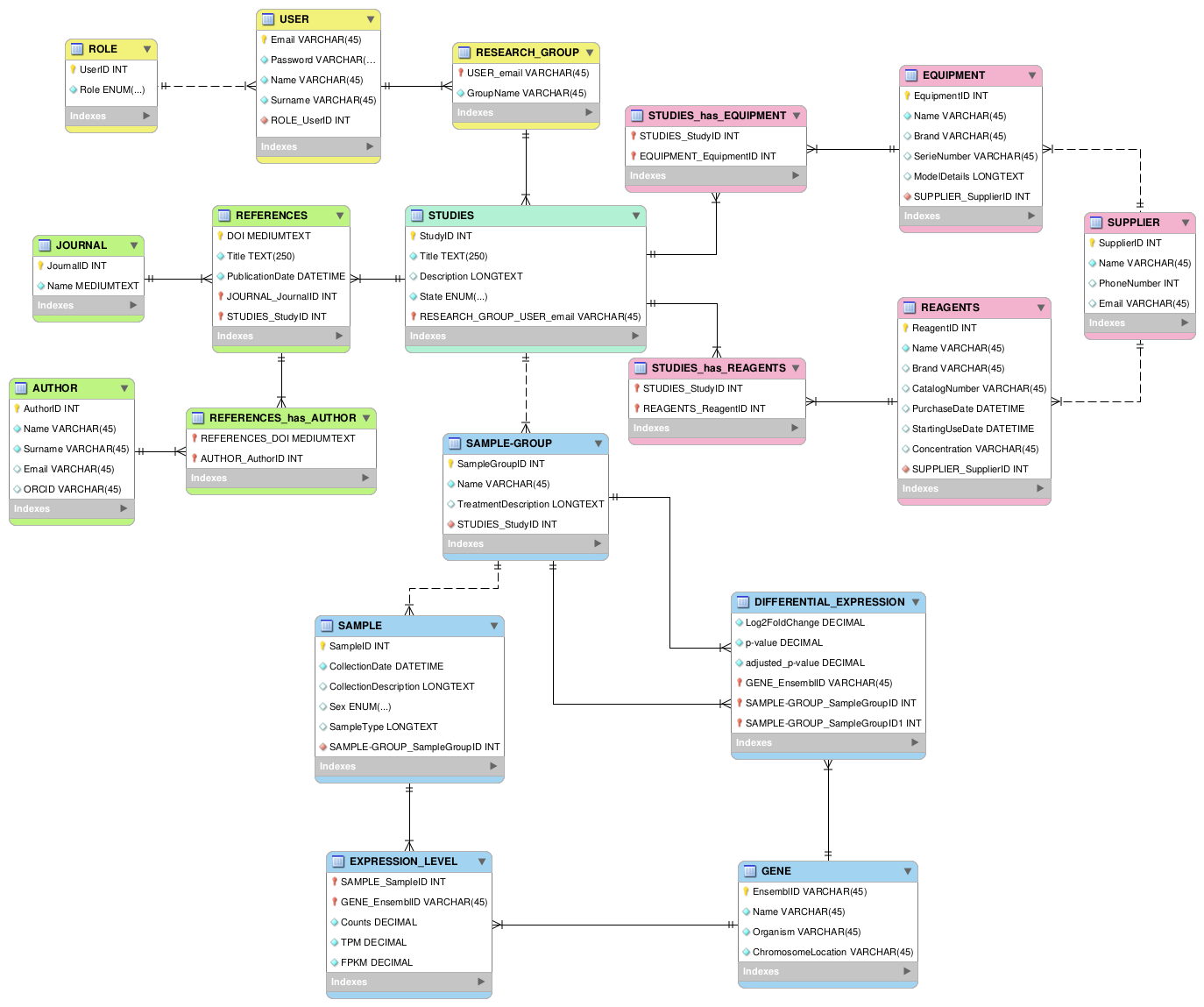
Understanding the Data Model
🟨 User Management
This section stores information related to users, roles, and research groups. Users can have different roles, such as researchers or technicians, and belong to research groups. The RESEARCH_GROUP entity allows multiple users to participate in different studies while maintaining structured participation.
🟪 Equipment & Reagents
This section manages the equipment and reagents used in the study. Each study can use multiple pieces of equipment and reagents, which are supplied by different vendors. The many-to-many relationship between STUDIES and EQUIPMENT/REAGENTS ensures that resources are properly tracked.
🟩 References & Publications
References are critical in research. This section stores scientific references, journals, and authors. Each reference can have multiple authors, and each study is linked to relevant scientific literature. The REFERENCES_has_AUTHOR entity manages this many-to-many relationship.
🟦 Experimental Data & Gene Expression
This section represents biological samples, gene expression values, and differential expression analysis. Each study can have multiple experimental groups (SAMPLE_GROUP), which contain multiple samples. Gene expression is recorded per sample, and values such as TPM, FPKM, and raw read counts are stored.
🔬 Differential Expression Analysis
Differential expression analysis compares gene expression between conditions. Since it involves two conditions, the DIFFERENTIAL_EXPRESSION entity links to two SAMPLE_GROUP entities, allowing for accurate comparisons of gene expression between groups.