Whole Genome Sequencing
Whole Genome Sequencing using SARS-CoV-2 as an example
Glossary
- Adaptors: Oligonucleotide that can be used for DNA barcode
- Amplicon: DNA amplified
- A-tailing: Enzimatic addition of adenines at -3’terminus, with purposes of sequencing.
- Barcoding: addition of known DNA sequence (barcode or index), to amplicon, allowing for parallel sequencing and for stratifying sample data informatically post/during sequencing.
- Consensus sequence: DNA, RNA or protein sequence generated from a set of aligned sequence calculated on the most frequent nucleic acids or residues in each position of the alignment.
- Library: in sequencing is a pool of DNA fragments attached to adaptors and/or other oligonucleotides used during sequence preparation.
- Trimming: removing ends of reads.
- Tagmentation: Initial step in library preparation, DNA is cleaved and tagged.
- Open reading frames:
Introduction to viral genomes
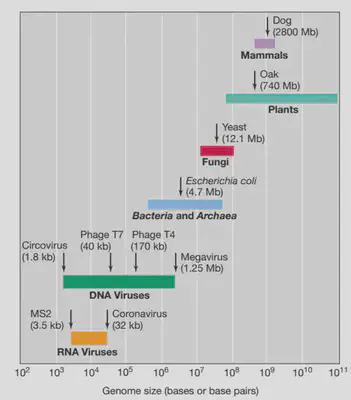
- RNA genomes tend to be smaller than DNA genome.
- General order of genome size: viral < prokaryote < eukaryote.
- In viral replication transcription is a mandatory step.
Baltimore Scheme for Viral Classification

- All RNA viruses are ss.
- All single-stranded DNA viruses are positive-strand viruses, with only one known exception, the Anelloviridae.
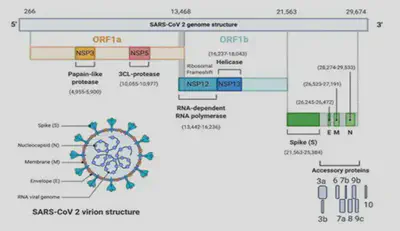
SARS-CoV-2 genome
- Coronaviruses (CoVs) are a group of enveloped ssRNA (+) viruses of 27-32kbp.
- Has at least 6 open reading frames (ORFs).
- The genes ORF1a and ORF1B encode 16 non-structural proteins (NSP1-16); which form a replication-transcription complex (RTC).
- The structural genes encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N).
Importance of viral sequencing
- Characterize viral genome
- Estimate variant’s prevalence
- Evaluate treatment effectiveness
- Understand evolution, epidemology diagnosis, surveillance
Bridge amplification
It was developed by Cambridge University’s Shankar Balasubramanian and David Klenerman, who later founded Solexa, which was acquired by Illumina. This method of sequencing DNA molecules of approximately 500 bp are used as substrates for repeated amplification synthesis reactions on a solid support (i.e., the flow cell) containing oligonucleotide sequences complementary to a ligated adapter.